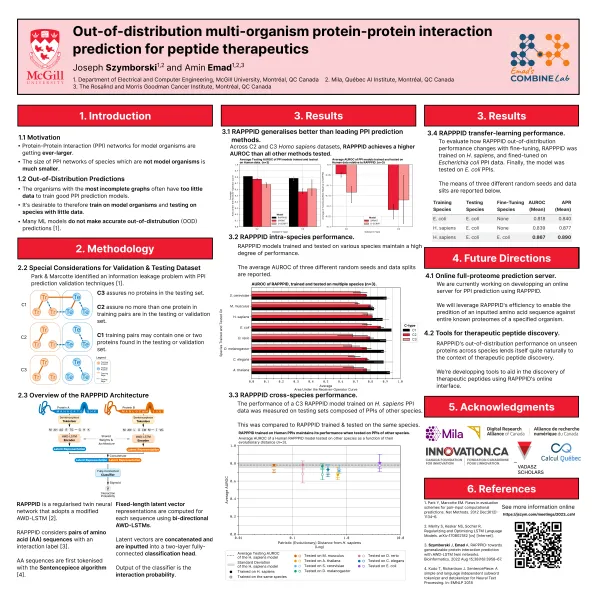
Poster
Download the poster we presented at CSHL 2023 here: ⇓ poster.pdf (1.6 MB)
Poster References
- Park Y, Marcotte EM. Flaws in evaluation schemes for pair-input computational predictions. Nat Methods. 2012 Dec;9(12):1134–6. DOI: 10.1038/nmeth.2259
- Merity S, Keskar NS, Socher R. Regularizing and Optimizing LSTM Language Models. arXiv:170802182 [cs] [Internet]. 2017 Aug 7 [cited 2021 Aug 10]; Available from: http://arxiv.org/abs/1708.02182
- Szymborski J, Emad A. RAPPPID: towards generalizable protein interaction prediction with AWD-LSTM twin networks. Bioinformatics. 2022 Aug 15;38(16):3958–67. DOI: 10.1093/bioinformatics/btac429
- Kudo T. Subword Regularization: Improving Neural Network Translation Models with Multiple Subword Candidates. arXiv:180410959 [cs] [Internet]. 2018 Apr 29 [cited 2021 Aug 10]; Available from: http://arxiv.org/abs/1804.10959
Code
You can access the latest code for RAPPPID on GitHub or cite our Zenodo repo:
Szymborski, Joseph, & Emad, Amin. (2022). Software for RAPPPID: Regularised Automative Prediction of Protein-Protein Interactions using Deep Learning (6d25213). Zenodo. DOI: 10.5281/zenodo.6709213
Data
You can download all the datasets used in the original RAPPPID paper on the Internet Archive, on Academic Torrents, or on Zenodo:
Szymborski, Joseph, & Emad, Amin. (2022). Data for RAPPPID: Towards Generalisable Protein Interaction Prediction with AWD-LSTM Twin Networks (Version 1) [Data set]. Zenodo. DOI: 10.5281/zenodo.6709790
Datasets used in the analysis for this poster will be released shortly.